Objective:
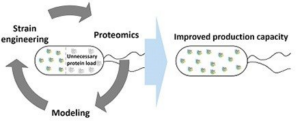
The main aim of LEANPROT is to develop an iterative systems biology-based strain engineering platform based on a novel approach of proteome optimization in iterative cycles of modelling and biological experiments which carries a substantial potential for addressing the aforementioned challenges in bioprocess development. That potential arises from the fact that cells express proteins unnecessary for growth under well-controlled optimal conditions, typically realized in biotechnological processes (e.g. flagellar, heat or acid stress proteins). This leads to non-efficient use of protein synthesis capacity (translation machinery) and energy for bioprocesses. Thus removing the expression burden of unnecessary proteins i.e. creation of lean-proteome strains, could enable to specifically manipulate the allocation of ribosomes for a higher synthesis of proteins leading to increased target molecule production.
Project summary:
Escherichia coli is a well-established and the most widely used organism for the production of recombinant proteins (used in medical and industrial applications, as molecular biology reagents, etc.). Production of proteins is the most resource exhaustive process for the cells and therefore needs to be optimized to achieve maximal productivities. The natural environment of E. coli is much harsher compared to the near optimal growth conditions used in production processes. In order to survive cells produce many native proteins that could be considered unnecessary for the cells in industrial production conditions. In this project, we aim to remove the most resource exhaustive unnecessary proteins from the host cells to free up resources for recombinant protein production. We will focus on this by using a novel metabolic modelling approach with constraints of protein production and cell geometry together with proteomics-based host cell physiology characterization. Novel lean-proteome strains with removed unnecessary proteins will be tested for the improved production capacity of several recombinant proteins used in research, industry and diagnostics.
Project No. ERASysAPP ID 2-12
Source of funding: FP7 EraSysApp (2nd call)
Project period: 01.12.2015 – 30.11.2018
Total budget: 1 624 000 EUR (210 000 for CSBG)
Project coordinator: Raivo Vilu, Competence Center of Food and Fermentation Technologies, Estonia
Project coordinator from our group: Senior researcher, Agris Pentjuss, Agris.Pentjuss@gmail.com
Results:
Stalidzans, E., Seiman, A., Peebo, K., Komasilovs, V., & Pentjuss, A. (2018). Model-based metabolism design: constraints for kinetic and stoichiometric models. Biochemical Society Transactions, BST20170263. https://doi.org/10.1042/BST20170263
Project partners:
- Technical University Berlin;
- Norwegian University of Science and Technology;
- Latvia University of Agriculture;
- Biotechrabbit;
- Competence Center of Food and Fermentation Technologies;